Imagine if there were ways for your mask to detect a COVID-19 infection or your bra to detect signs of breast cancer. Researchers at Tufts University and the University of Washington are working to make these speculations a reality. Labs at the two universities recently developed a novel way to detect viruses, toxins and other biomarkers through the use of de novo protein switches in a silk fibroin matrix. The research, published in Advanced Materials in December 2022, stemmed from a collaboration between the Silklab at Tufts and the Institute for Protein Design at the University of Washington.
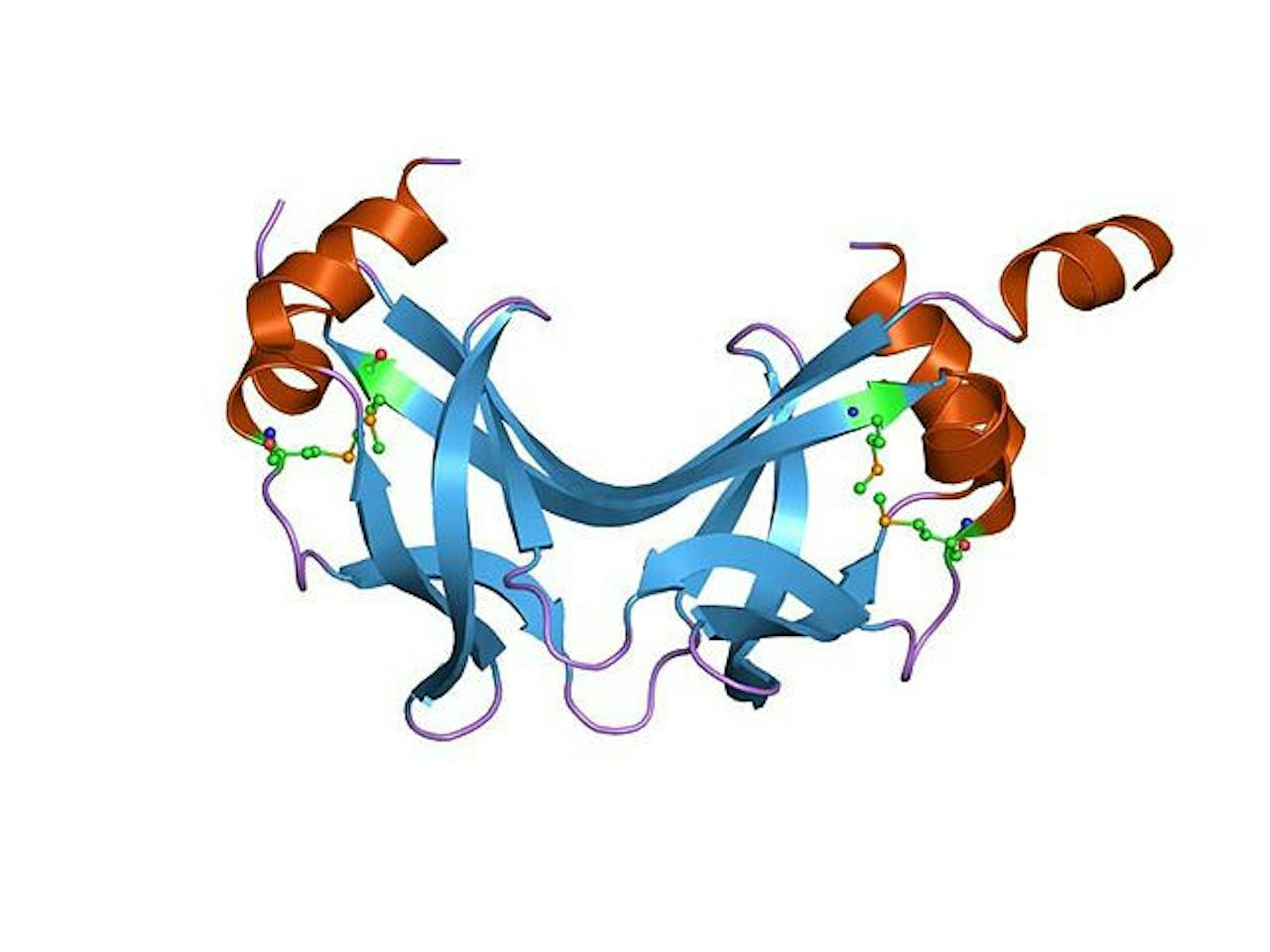
Alfredo Quijano Rubio, a former graduate student at the University of Washington and a co-author of the study, explained that a protein switch is a computationally designed protein molecule that can open and close depending on the external stimuli present. In the context of this research, these protein switches are used as biosensors, opening and closing to detect the presence and amount of a particular analyte.
“The key part is that this is very different from your classic biosensors, which are usually based on antibodies and require multiple steps,” Quijano Rubio said. “[The protein switch mechanism] all happens in solution because the proteins have been designed to open and close in solution. It’s basically a one-step reaction.”
To test the specificity of these protein switches for various molecules, the researchers looked at the receptor binding domain of the SARS-CoV-2 spike protein; the anti-hepatitis-B antibody; the human epidermal growth factor receptor 2, which is implicated in breast cancer detection; and Botulinum neurotoxin B, a protein that can be found in spoiled foods. Quijano Rubio said that the protein switches are pretty sensitive for these specific molecules and can detect concentrations in the low picomolar range.
Fiorenzo Omenetto, a professor in the Department of Biomedical Engineering, is the primary investigator and director of the Silklab and a co-author of the protein switches study. He said this collaboration combines the University of Washington’s protein switch technology and Tufts’ silk fibroin matrix development. The research process resulting in these findings lasted about two years.
“It seemed like it was an opportunity to have a material that combined these things, and … the result would have been greater than its individual parts,” Omenetto said.
Omenetto recalled the day he met David Baker, head of the Institute for Protein Design and another co-author on the study. The two connected almost immediately over their respective projects.
“The real story is that we were both kind of sitting away from the crowds in a very self-promotion-heavy … conference, with a lot of ‘alpha types’ telling you how they were going to solve the problems of the world,” Omenetto said. “Then we went and sat in a quiet space and then I said, ‘I liked what you commented,’ and he said, ‘I liked what you commented,’ … and we said, ‘We should think of doing something together.’”
Quijano Rubio elaborated that the protein switch is made up of two parts. The platform of the protein remains constant while the binding domain is modified based on the specific molecule it is trying to detect. Once created, the biosensor is combined with the silk matrix. The protein switches made at the University of Washington are then shipped to the lab at Tufts to be incorporated into the silk matrix.
Omenetto explained that the silk fibroin matrix is especially useful because it’s versatile, implantable and even food safe. Different biomaterials were tested throughout the process, but silk was chosen because it worked so well as a water-based stabilizer for the protein switches.
“There’s a combination of really favorable features that silk has, from the mechanical standpoint, from the transformation standpoint, in all these different materials,” Omenetto said. “If you had to single out the one thing that is the main difference, [it’s that] the processing of silk into all of these different formats starts from a water-based solution. That’s actually very important for biologicals [and] for foods.”
An analyte is sensed and visualized in solution through a Key-Cage mechanism, as described in the paper. Quijano Rubio explained that this is a two-component system in a state of equilibrium with the analyte. The cage portion contains the binding domain for the molecule as well as a luciferase component, which creates fluorescence in the presence of a desired molecule. The only way to form the luminescent complex and detect the molecule of interest is to have both the key and the analyte present to ‘unlock’ the cage and allow for the luciferase reaction to take place. The amount of light visualized is proportional to the amount of analyte present.
Omenetto said that this technology is applicable outside of the laboratory in so many ways, as long as there are passionate people eager to work on developing the materials and scaling them up. He mentioned that he could see protein switch technology applied to materials that detect COVID-19, sense breast cancer and monitor mental health through chemical indicators implicated in mental illnesses.
“I think there’s a lot of good that can be done and a lot of challenges too … [that] are not going to be trivial,” Omenetto said. “It’s a matter of finding people that are as invested and as passionate and committed as you to doing these things. … These people are out there, but you have to meet, you have to cross paths.”